Recent tissue-level observations made indirectly via flow cytometry show that endoreplication (partial or complete duplication of the nuclear genome in the absence of subsequent cell division) may represent a significant component of a plant’s developmental programme. Bateman et al. directly observe ploidy variation among cells within individual petals, relating size of nucleus to cell micromorphology and (more speculatively) to function.
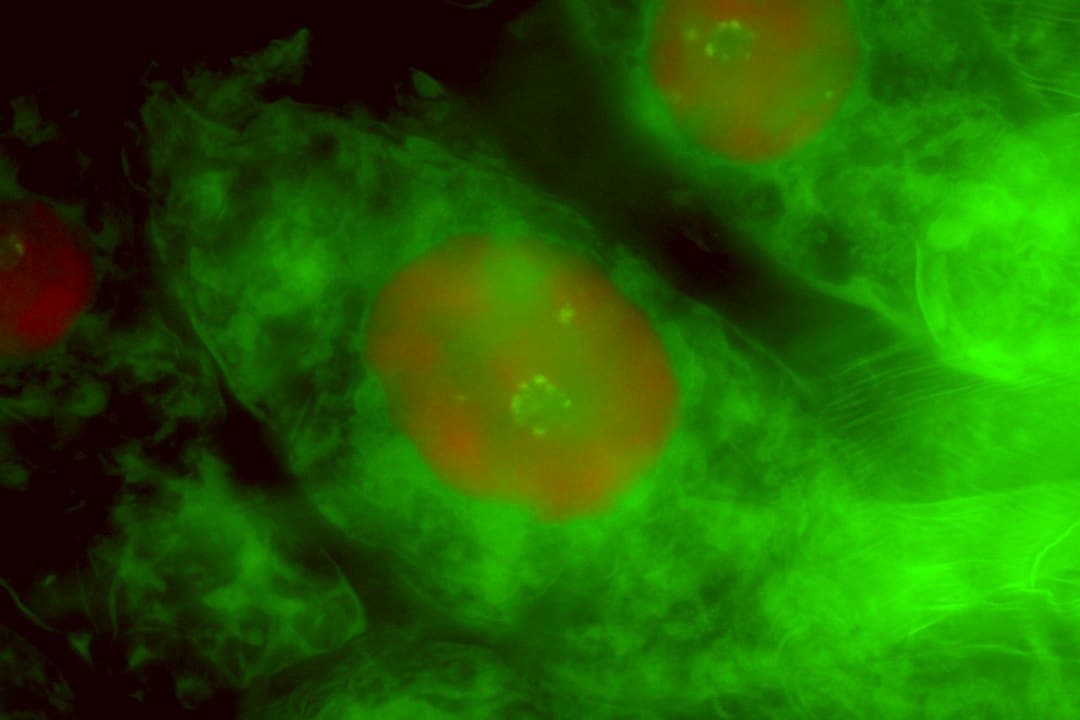
The authors compared the labella (specialized pollinator-attracting petals) of two European orchid genera: Dactylorhiza has a known predisposition to organismal polyploidy, whereas Ophrys exhibits exceptionally complex epidermal patterning that aids pseudocopulatory pollination. Confocal microscopy using multiple staining techniques allowed the team to observe directly both the sizes and the internal structures of individual nuclei across each labellum, while flow cytometry was used to test for progressively partial endoreplication. Endoreplication involved repeated near-doubling of the genome and proved especially frequent in the more complex labellum of Ophrys, reaching 16C polytene nuclei in large trichomes.
Bateman et al. find that in Dactylorhiza, endoreplication was comparatively infrequent, reached only low levels, and appeared randomly located across the labellum, whereas in Ophrys endoreplication was commonplace, being most frequent in large peripheral trichomes. Endoreplicated nuclei reflected both endomitosis and endocycling, the latter reaching the third round of genome doubling (16C) to generate polytene nuclei. All Ophrys individuals studied exhibited progressively partial endoreplication.




