A team from the University of Illinois has developed a modeling framework connecting enzyme activity related to dynamic photosynthesis with yield for the first time.
“A previous model combined data from the genetic, metabolic, and leaf levels to simulate photosynthesis over time. We took it further and connected the metabolic level with the whole canopy and simulated yield over a growing season,” said Yufeng He, a postdoctoral researcher in the Matthews Group at Illinois. “The new model allows us to examine how changes in enzyme activities in response to the environment can affect yield by connecting the environmental variation experienced by the crops in the field to the metabolic processes.”
In a recent study, published in in silico Plants, He and others used their new model to demonstrate that increasing the concentration of key enzymes affecting photosynthesis could boost yield. Previously, scientists simulating the enzyme activity of photosynthesis were limited to studying the photosynthetic response at the leaf-level, ignoring the plants’ dynamic physiological responses to environmental factors like light, temperature, and water that have an integrated impact over a growing season. For anyone who has been in a crop field, that is very unrealistic.
“Plants don’t exist in a stable environment. We can use this work to study the sensitivity of enzymes under the different environmental conditions that plants experience,” said Megan Matthews, assistant professor in civil and environmental engineering at Illinois. “The model will allow us to see which photosynthetic enzymes are limiting in different environments, and how they can lead to yield gain under future climatic conditions.”
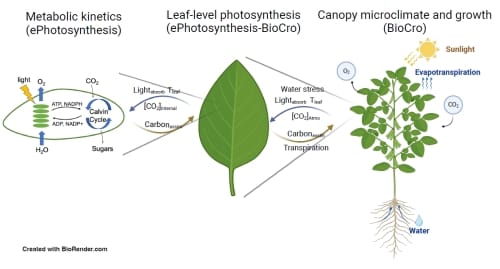
Using a dynamic model allows researchers to more accurately predict how plants will respond in their changing environment. Throughout a normal day, plant canopies experience significant fluctuations in the photosynthetic process due to factors such as cloud cover and shading from other plants. Incorporating this variability into the model enhances its accuracy and reliability. In addition, identifying which enzymes are limiting plant photosynthesis and growth in various environments like drought, heavy rain, or elevated CO2 is crucial. This can enable plant breeders and engineers to target specific enzymes for crop improvement. By optimizing enzyme activity, it is possible to develop plant varieties that are more resilient and productive, ultimately leading to increased yields.
He and Matthews hope that their model, available open source on Github, can help researchers better understand and predict how specific photosynthetic enzymes affect crop growth and yield in different environments. This work is part of Realizing Increased Photosynthetic Efficiency (RIPE), an international research project engineering crops to be more productive by improving photosynthesis, the natural process all plants use to convert sunlight into energy and yields.
READ THE ARTICLE:
Yufeng He, Yu Wang, Douglas Friedel, Meagan Lang, Megan L Matthews, Connecting detailed photosynthetic kinetics to crop growth and yield: a coupled modelling framework, in silico Plants, Volume 6, Issue 2, 2024, diae009, https://doi.org/10.1093/insilicoplants/diae009





